Using simulated CBCT images in deep learning methods for real CBCT segmentation
PO-1646
Abstract
Using simulated CBCT images in deep learning methods for real CBCT segmentation
Authors: Nelly Abbani1, Franklin Okoli2, Vincent Jaouen2, Julien Bert2, David Sarrut1
1CREATIS, UMR 5220, Lyon, France; 2LaTIM, UMR 1101, Brest, France
Show Affiliations
Hide Affiliations
Purpose or Objective
Segmenting organs in Cone-Beam CT (CBCT) images would allow to adapt the dose delivered based on the organ deformations that occured between the treatment fractions. However, this is a difficult task because of the relative lack of contrast in CBCT images, leading to high inter-observer variability. Deep-learning based automatic segmentation approaches have shown impressive successes and may be of interest here but required to train a convolutional neural network (CNN) from a database of segmented CBCT images, which can be difficult to obtain. In this work, we propose to train a CNN from a database of artificial CBCT images simulated from planning CT for which it is easier to obtain the organ delineations.
Material and Methods
Simulated-CBCT (sCBCT) images were computed from readily available segmented planning CT images, using the GATE Monte Carlo [Sarrut2014] simulation toolkit. The simulation reproduces the CBCT acquisition process of an Elekta XVI device, tracking the beam of particles from the x-ray source, through the CT images and in the flat panel detector. The reference delineations in the CT images, performed during planning by the clinical team, were then projected on the sCBCT images, resulting in a database of segmented images that could be used to train a CNN. The studied segmentation labels were: bladder, rectum, and prostate contours. The CNN is a 3D-Unet called nnUnet, with a batch size of 2, patch size of (191,257,219), and 2-mm target spacing. The training is run over 200 epochs. The optimizer used is stochastic gradient descent. Several models were trained: 1) on 138 real CBCT images delineated by experts from 7 patients, 2) on 90 sCBCT images from 90 patients. The evaluation was performed on a different dataset of segmented CBCT by comparing predicted segmentation with reference ones thanks to Dice similarity score and 95th percentile of Hausdorff distance.
Results
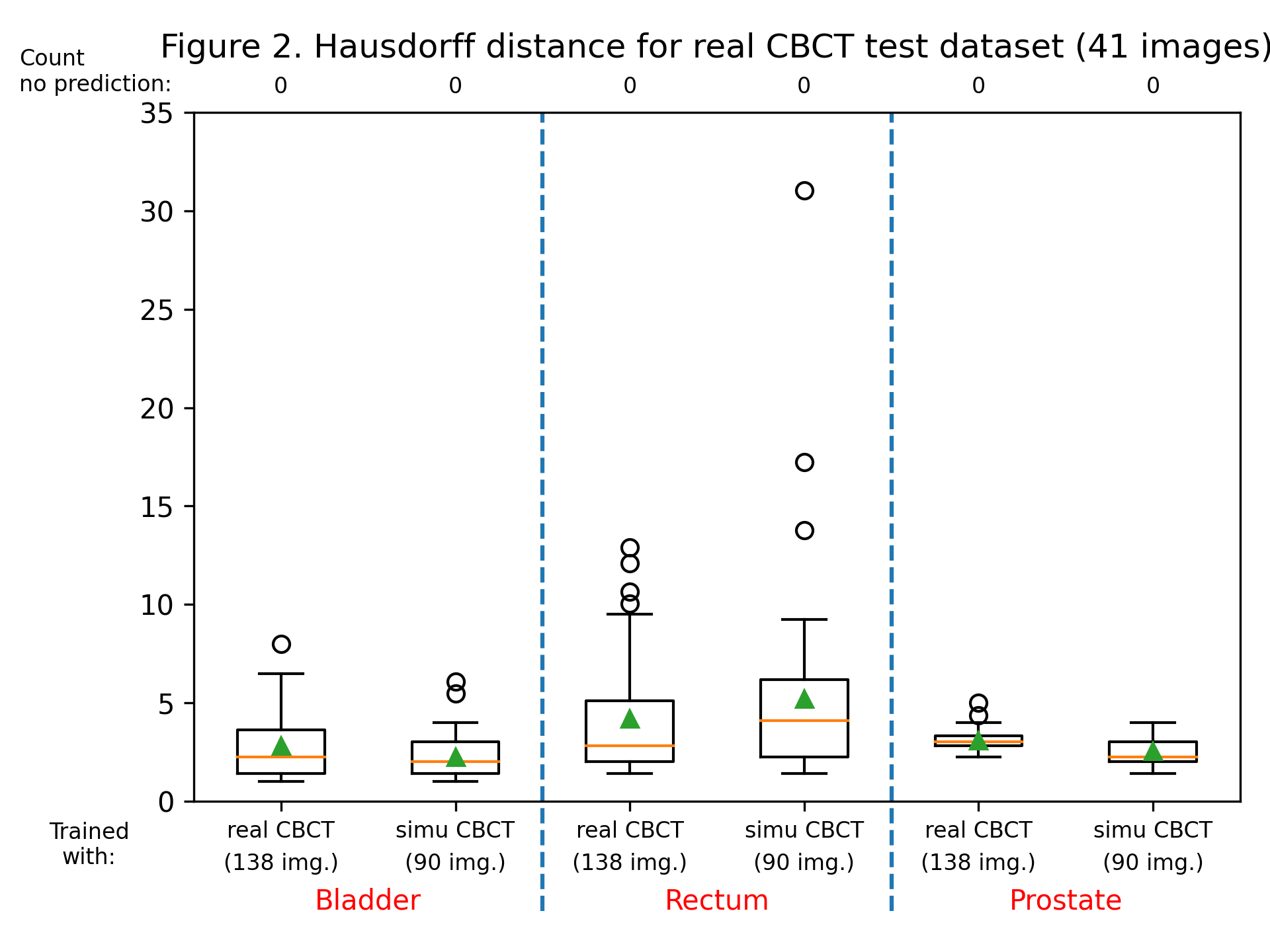
The dice score and Hausdorff distance were calculated for each label, for each patient, for both trained models. Dice scores are summarized in figure 1, and the Hausdorff distance in figure 2. Training with sCBCT images was found to lead to comparable results than using real CBCT images (an average dice scores of around 0.9 for the bladder label, 0.8 for the rectum and prostate label, and an average Hausdorff distance between 2 and 5 mm for all labels). Although the number of real CBCT images (138) is higher than those of sCBCT, they have less variability as they come only from 7 patients, explaining the slightly lower performance.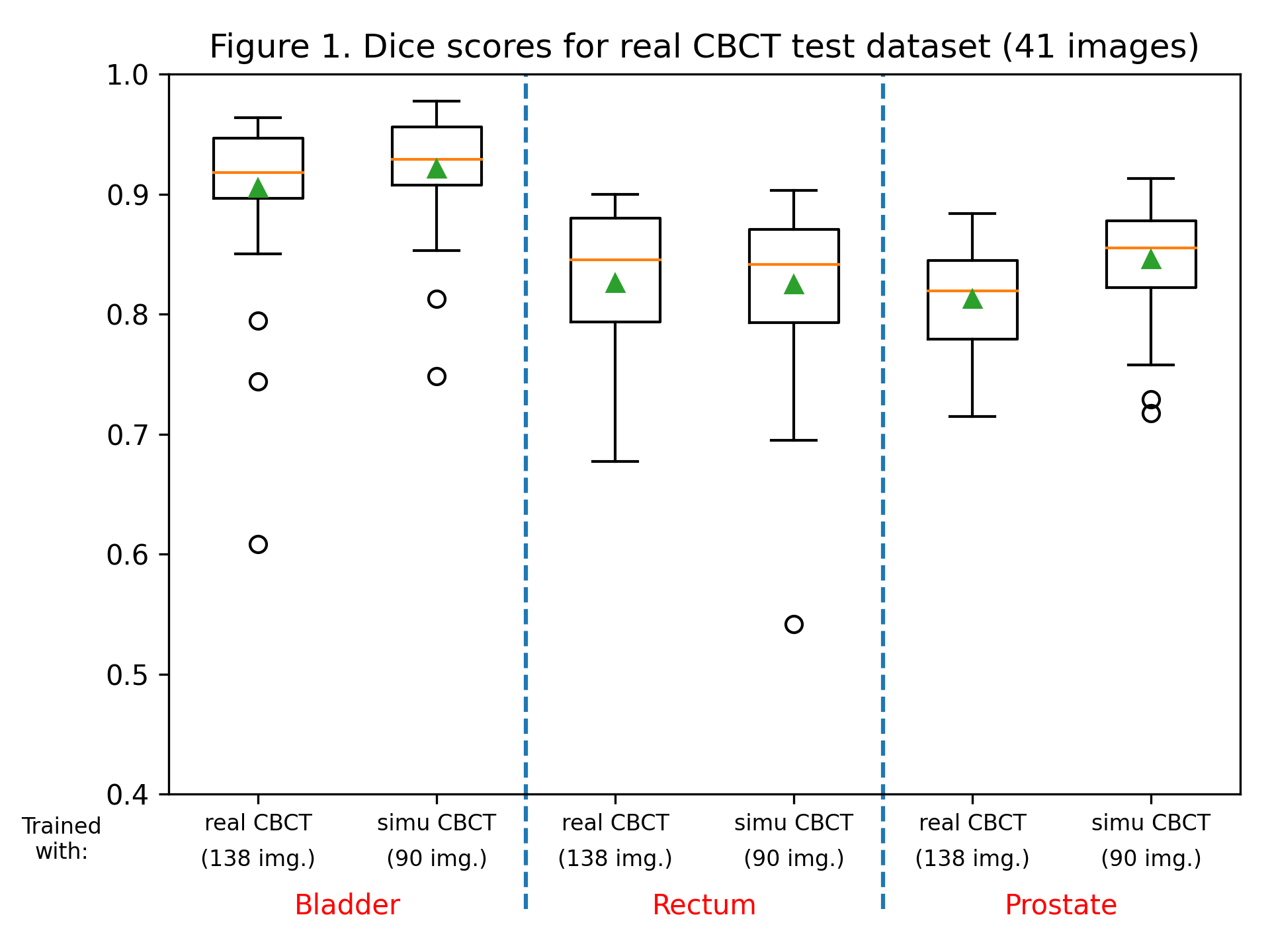
Conclusion
We proposed to use simulated CBCT images to train a CNN segmentation model avoiding the need to gather complex and time-consuming reference delineations on CBCT images. As a perspective, robustness of the method against datasets from different institutions and from different CBCT devices will be assessed.