Three
lung cancer patient cohorts and various EPID dosimetry modalities were
considered: (1) 69 stereotactic body radiotherapy (SBRT) plans with 2D
time-integrated (TI) pre-treatment EPID dosimetry, (2) 47 VMAT or hybrid plans
(with both static beams and VMAT arcs) with 2D-TI transit EPID dosimetry and
(3) 46 VMAT plans with 2D time-resolved (TR) transit EPID dosimetry. Clinically
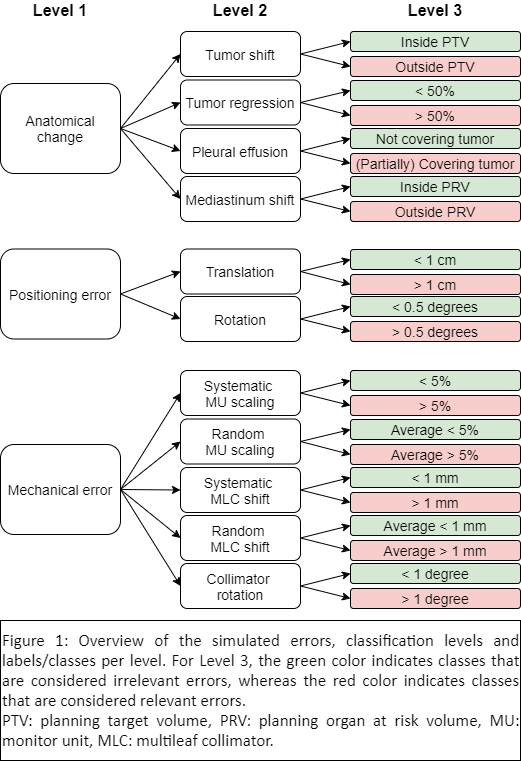
realistic ranges of mechanical errors (all cohorts), anatomical changes (cohort
2 and 3) and positioning errors (cohort 2 and 3) were simulated (Figure 1). Predicted
portal dose images (PDIs) containing errors were compared to error-free PDIs
using (3%, 3mm) gamma analysis. For cohort 1, (3%, 1mm) gamma analysis was
additionally performed and evaluated. CNNs were optimized and trained to
classify errors using gamma maps as input. Three classification levels were
assessed (Figure 1), from coarser to more detailed: Level 1 (main error type,
e.g., anatomical change or mechanical error – cohorts 2 and 3), Level 2 (error
subtype, e.g., tumor regression or systematic MLC shift – all cohorts) and
Level 3 (error magnitude, e.g., >50% tumor regression or MLC shift > 1 mm
– all cohorts).